Open-Source
Our software is
Free software
Dlab software:
OSPREY
DISCO
PIV
LibProtNMR
ConDIOs
RDC-PANDA
NASCA
POOL
Q5
CRANS
Software previously released by the Donald Lab:
Exact-2NH
MTC
NVR/HD/GD
Rage/Enrage
Software Developed by the Donald Lab
Protein Interaction Viewer
Download PIV | License | Contact
Welcome to the website for the Protein Interaction Viewer software.
About PIV (Protein Interaction Viewer)
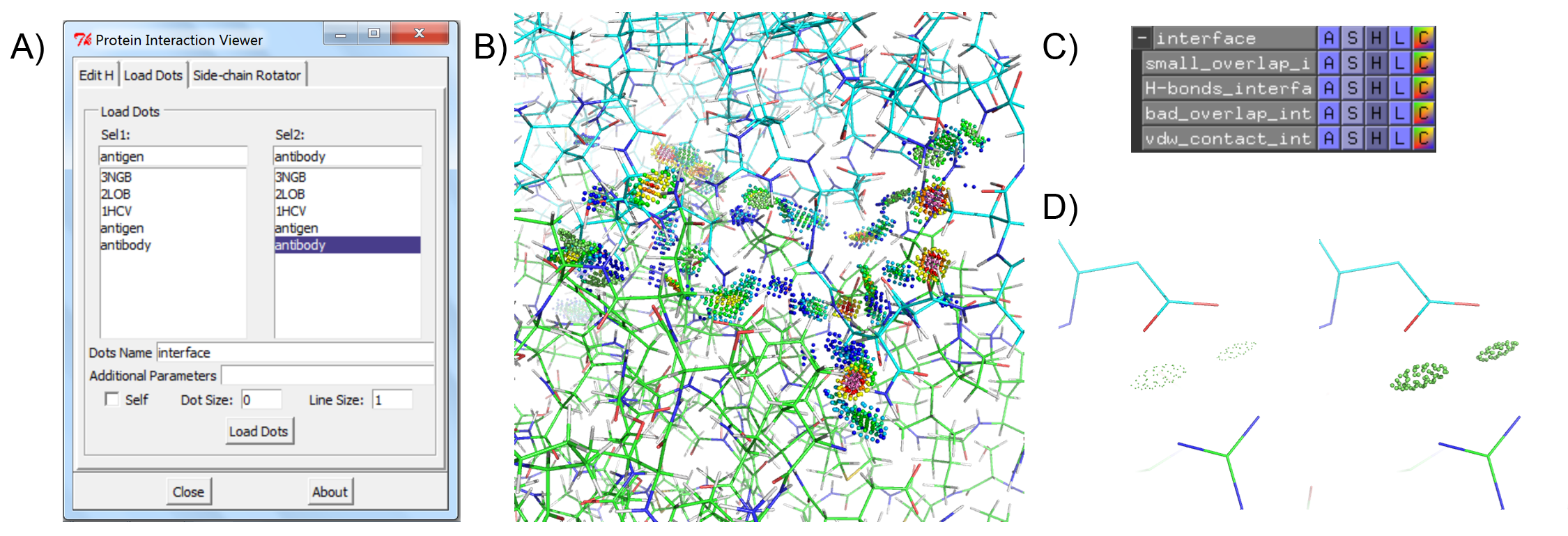
The Richardson Lab has developed a suite of tools for validating protein structure that is made available through the MolProbity server. Of particular interest for visualizing protein interactions is the Probe program, which generates contact dots between protein residues that are close enough to interact with one another. The contact dots visually represent favorable van der Waals interactions, steric clashes, and hydrogen bonding. These visual aids are useful to rapidly identify both favorable interactions and problems with the protein structure, but are only natively available in a limited number of molecular visualization programs. Protein Interaction Viewer is a PyMOL plugin that allows Probe contact dots to be visualized within PyMOL, a widely-used molecular viewer.
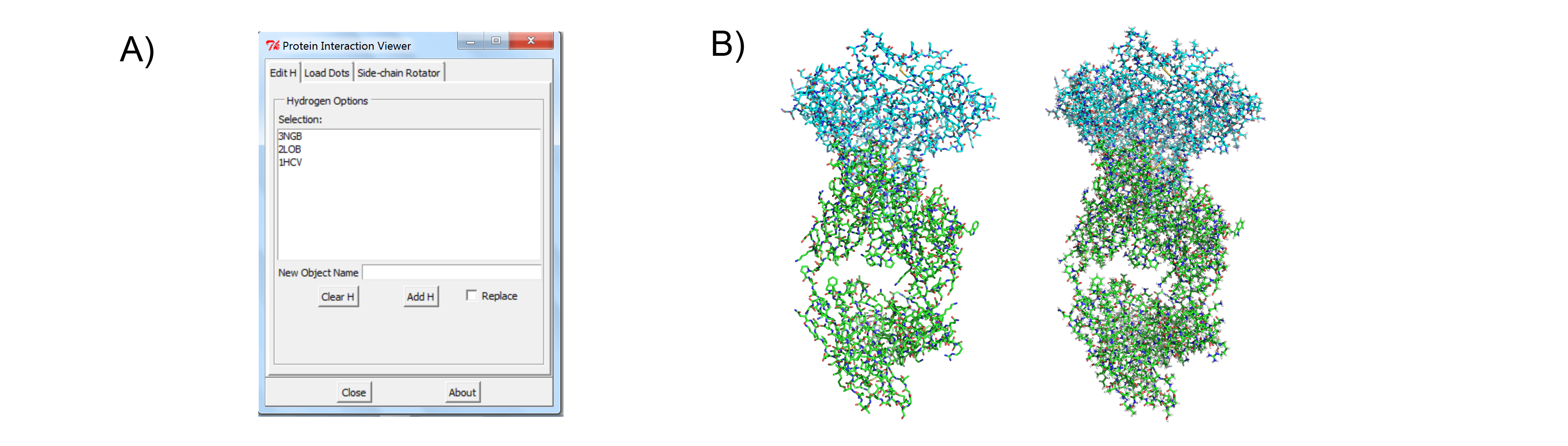
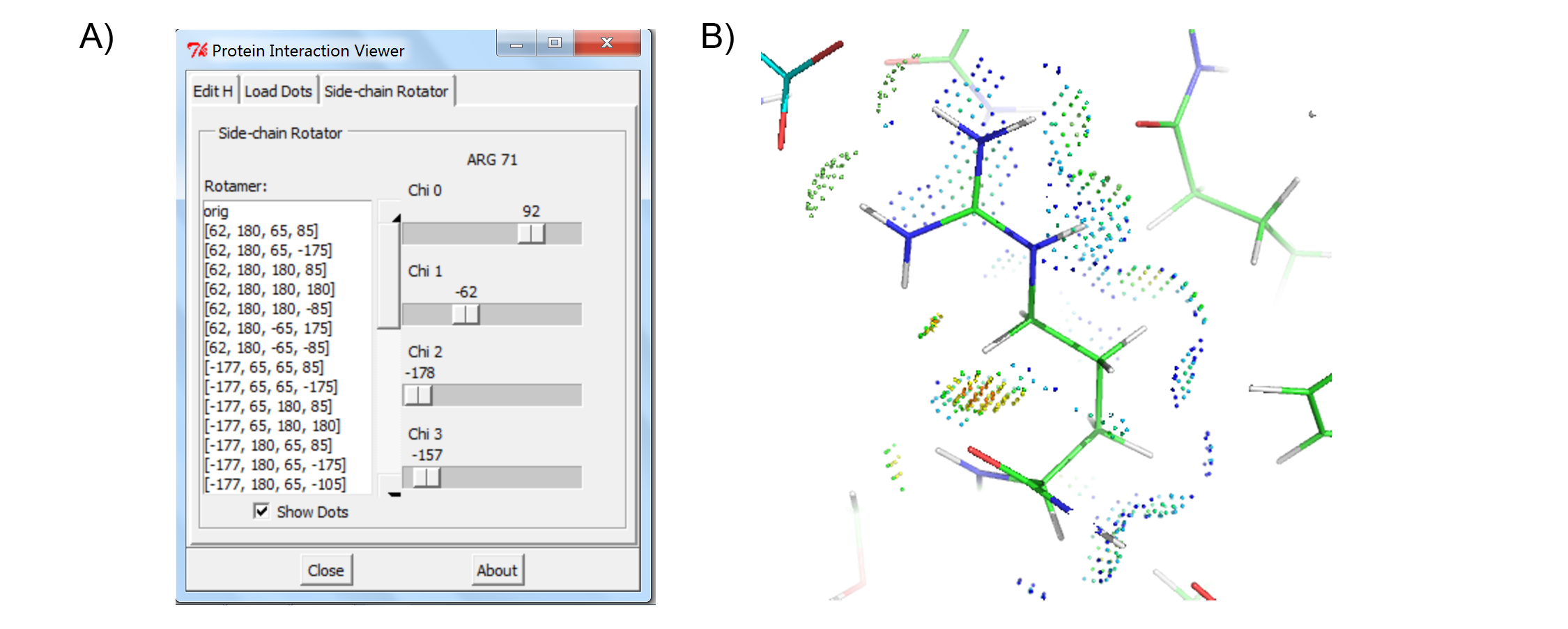
The Protein Interaction Viewer (PIV) PyMOL plugin can carry out three main functions. First, to properly calculate molecular interactions within a protein structure the protein must be fully protonated. PIV provides easy access to the program Reduce that can either add or remove hydrogens from a PDB structure that has been loaded into PyMOL (Figure 1). Second, PIV allows the user to create and visualize contact dots for a single PyMOL selection or between two PyMOL selections (Figure 2). The dots are created as PyMOL CGO objects, one object for each type of contact (i.e. small overlap, H-bond, bad overlap, and vdW interaction). The four objects are all added to the same PyMOL group for convenience. Finally, PIV allows the user to easily rotate any side chain within the protein (Figure 3). While the user is rotating a side-chain, they can enable contact dots between that side-chain and the rest of the protein.
Installation & Usage
Please see Protein Interaction Viewer's Github repository for installation and usage instructions.
Citation
See License
