The translation-only RMSD (TRMSD) was computed between the model
oriented
according to each cross-rotation peak and each chain in the crystal
structure. We define the
TRMSD of two proteins to be the minimum main chain Ca RMSD achievable when
the molecules
are only allowed to translate relative to one another (i.e., rotations are
not allowed). Using this
measure we are able to quantify the similarity between models oriented by
each cross-rotation peak
and each monomer of the crystal structure.
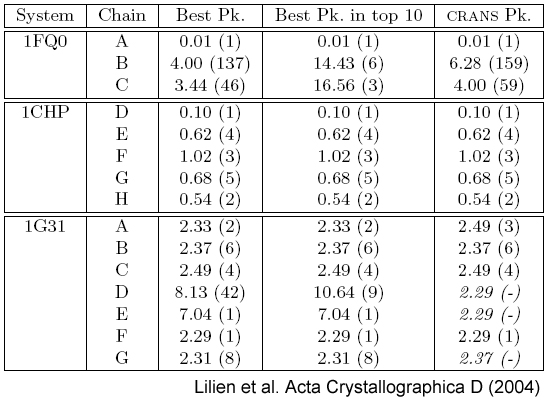
The table displays the
TRMSDs measured for a CRANS analysis for the three test systems, 1FQ0, 1CHP, and 1G31. Column 2 lists the
PDB chain identifier used in computing the TRMSD for the specified row. The ‘Best Pk.’ column lists
the minimum TRMSD observed between the specified chain and all cross-rotation peaks along with its
corresponding cross-rotation peak index (in parentheses). Cross-rotation
peaks are ordered by sorting them
based on the cross-rotation function score where a lower index corresponds
to a higher (better) cross-rotation
score. The TRMSD and peak index of the peak with the smallest TRMSD among
the top 10 cross-rotation
function ranked peaks is listed in the ‘Best Pk. in top 10’ column. The
TRMSD of the CRANS identified
peak of the NCS-consistent rotation set corresponding to the specified
chain and its cross-rotation peak
index (in parentheses) is listed in column ‘CRANS Pk.’. Peaks computed by
CRANS (i.e., those missing in
the cross-rotation peak list) are shown in italics with a dash for the peak
index. |